Research in Progress awardees
1st Place
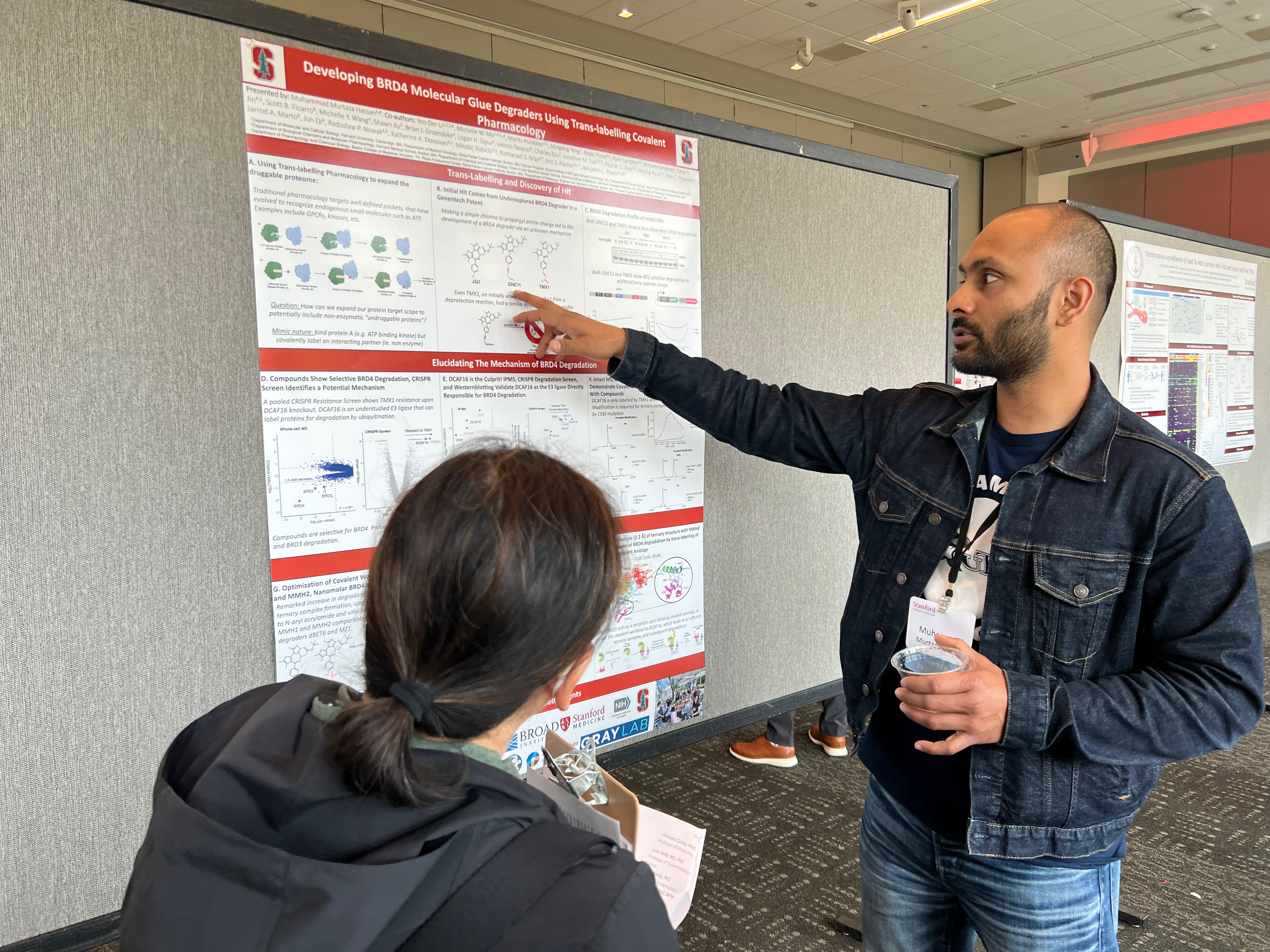
Title: Developing BRD4 molecular glue degraders using trans-labelling covalent pharmacology
PI(s): Nathaniel Gray, PhD
Presenter(s): Muhammad Murtaza Hassan, PhD
Abstract: Expanding the druggable proteome is of great interest in the drug discovery paradigm. Non enzymatic targets however, lack well-defined chemical recognition pockets and are hard to chemically target since non-enzymatic protein functions such as protein-protein interactions rely on shallow grooves and largely surface-surface interactions. However, due to the advancements in targeting enzymatic proteins and developments in unique chemical designs, it may be possible to design a drug that binds and enzymatic protein, which then delivers the drug to an interacting protein partner. In the context of covalent pharmacology, a covalent compound could presumable bind a protein target (A) but covalently target it's interacting protein partner (B), which we describe as 'trans-labelling'. Herein, we describe the covalent derivatization of JQ1 (compounds MMH1 and MMH2), a BRD4 inhibitor, to 'trans-label' DCAF16, an E3 ligase responsible for ubiquitination and subsequent protein degradation of its substrates. This trans-labelling event then leads to the DCAF16 forming a covalent adduct with MMH1 and MMH2, leading to subsequent robust degradation of BRD4. This study illustrates the characterization of the first trans-labelling covalent-pharmacology in literature, and demonstrates its potential for achieving unique pharmacological consequences such as protein degradation.
2nd Place
Title: Noninvasive surveillance of head & neck cancers with viral and tumor cell-free DNA
PI(s): Ash Alizadeh, MD, PhD; Dimitri Colevas, MD
Presenter(s): Joe Schroers-Martin MD, Andrea Garofalo BS, Steven Tai Lai BA, Elizabeth Winters MS, Savanna Biedermann BS, Saad A. Khan MD, Maximilian Diehn MD, PhD, A. Dimitrios Colevas MD, Ash Alizadeh MD, PhD
Abstract: Head and neck cancers are associated with chronic viral infections including human papillomavirus (HPV) and Epstein-Barr virus (EBV). Cell-free DNA (cfDNA) plays an increasing role in the detection of cancers. We therefore explored whether viral and tumor-derived cfDNA are detectable in the blood and urine of head and neck cancer patients. We prospectively enrolled 13 pre-treatment patients with virus-positive localized or locally advanced disease, including 3 EBV+ patients (2 NPC, SCC) and 10 HPV+. Post-treatment sample collection is ongoing. In a pilot cohort we analyzed 17 urine and 15 plasma samples from 13 patients, including 6 post-treatment (urine=4, plasma=2). Somatic coding mutations in genes including TP53, FAT1, PIK3CA, KMT2D, and NSD1 were detected in pre-treatment plasma samples at a mAF of 1.4%. Concordant mutations were detected in 9/13 paired pre-treatment urine samples (69%), but at median 5-fold lower AF.
Viral DNA corresponding to tumor pathology was detected in all plasma (100%) and 11/13 urine samples (84%), but at much lower levels in urine (median -3.9 log). Viral levels dropped with therapy in 5/6 post treatment samples. While plasma was significantly enriched in HPV16, urine specimens were enriched in JC and BK polyomavirus. Overall, our data suggests joint hybrid capture enrichment of viral and tumor cfDNA as a promising strategy in head & neck cancers, with higher concentrations observed in plasma.
Shared Resource awardee
Title: The Human Immune Monitoring Center at Stanford: Revealing the Human Immune System in Cancer
Presenter(s): Caroline Duault, PhD; Holden Maecker, PhD
Abstract: The Human Immune Monitoring Center (HIMC) provides standardized, state-of-the-art immune monitoring assays at the DNA/RNA, protein and cellular level. We are also dedicated to continuously improve our expertise and to participate in the development of new technologies to push back the limits of Science. We currently offer multiplex Luminex and Olink for human and mouse samples; RNA/DNA extraction from blood, FFPE tissue and cell-free plasma; genomic assays, including BD Rhapsody single-cell transcriptomics and proteomics, microfluidic qPCR arrays as well as single-cell targeted RNAseq for T cells; and flow cytometry and CyTOF with immunophenotyping, intracellular cytokine and phospho-flow panels.
As part of our recent projects, we used flow cytometry and CyTOF to delineate the phenotypic and functional characteristics of the NK cell subsets in the bone marrow and peripheral blood of B/T Acute Lymphoblastic Leukemia (ALL) patients. We showed that NK cells in B/T-ALL display chronically hyper-activated phenotype with increased production of cytokines, eventually leading to exhaustion and failure to kill target cells. Moreover, using the immune signature we defined, we demonstrated that activated NK cells predict poor clinical prognosis in high-risk B/T-ALL. This is an example among many of a successful collaboration between our lab and others, ranging from Stanford University, to other academic institutions or industry.